Nature Communications (2022)
Arbuscular mycorrhiza (AM) is a widespread symbiosis between roots of the majority of land plants and Glomeromycotina fungi. AM is important for ecosystem health and functioning as the fungi critically support plant performance by providing essential mineral nutrients, particularly the poorly accessible phosphate, in exchange for organic carbon.
AM fungi colonize the inside of roots and this is promoted at low but inhibited at high plant phosphate status, while the mechanistic basis for this phosphate-dependence remained obscure. Here it is demonstrated that a major transcriptional regulator of phosphate starvation responses in rice PHOSPHATE STARVATION RESPONSE 2 (PHR2) regulates AM. Root colonization of phr2 mutants is drastically reduced, and PHR2 is required for root colonization, mycorrhizal phosphate uptake, and yield increase in field soil. PHR2 promotes AM by targeting genes required for pre-contact signaling, root colonization, and AM function. Thus, this important symbiosis is directly wired to the PHR2-controlled plant phosphate starvation response.
Keywords: arbuscular mycorrhizae, endomycorrhizae, phosphorus, organic agriculture, transcriptional factor (PHR2)
https://www.nature.com/articles/s41467-022-27976-8
Photo:
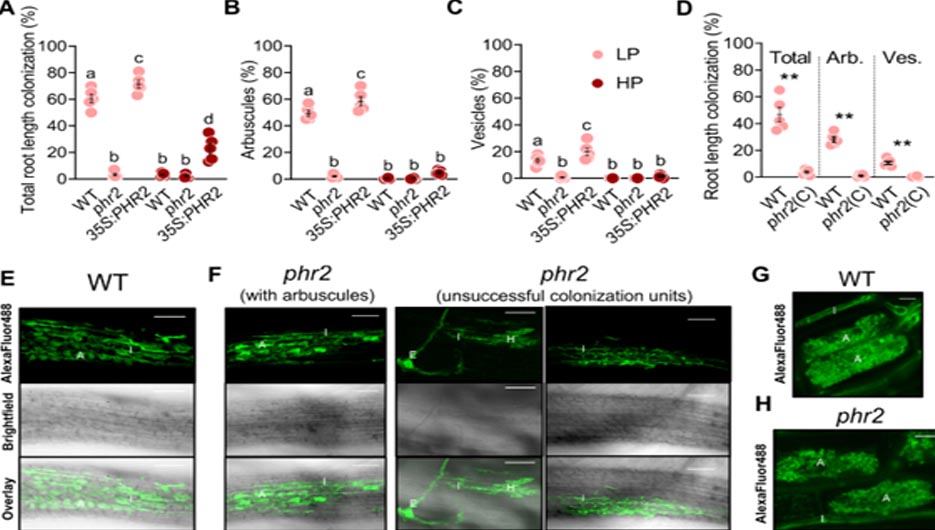
Percent root length colonization (RLC) total (A), arbuscules (B), and vesicles (C) of the indicated genotypes inoculated with R. irregularis (AMF) for 7 weeks at low phosphate (25 µM Pi, LP) or high phosphate (500 µM Pi, HP). D Percent RLC of the indicated genotypes inoculated with AMF for 7 weeks at LP. Arb.: Arbuscules; Ves.: Vesicles. Confocal images of AMF in roots of wild type (E) and phr2 (F). Arbuscule phenotype in wild type (G) and phr2 (H) roots. E, extraradical hypha; H, hyphopodium; I, intraradical hypha; A, arbuscule. Scale-bars: E, F: 100 μm and G, H: 10 μm. Statistics: Individual data points and mean ± SE are shown. A–C N = 5 independent plants; Brown-Forsythe and Welch’s One-Way ANOVA test with Games–Howell’s multiple comparisons test. Different letters indicate statistical differences. D N = 5 independent plants; Mann–Whitney test (two-tailed) between phr2(C) and wild type for Total (p = 0.0079), arbuscules (p = 0.0079) and vesicles (p = 0.0079). Asterisks indicate significance of difference: ** p ≤ 0.01. E–H The phenotype was observed in 11 (6 + 5) independent plants in two independent experiments.


